the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Genetic diversity and differentiation of invasive Acacia longifolia in Portugal
Sara Vicente
Cristina Máguas
Helena Trindade
Acacia longifolia is an aggressive invader worldwide. This species was brought to Portugal by the forestry services and is now found throughout the country with very strong ecological and social impacts. Although several ecological and physiological studies have been carried out, molecular studies in this species are sparse. Particularly, genetic variability evaluation in invasive ranges clearly deserves more attention. The aim of this study was to evaluate the genetic diversity and the genetic differentiation among populations of the alien invasive A. longifolia under different Mediterranean conditions. For that we studied three sandy dune populations along the Portuguese coast: Osso da Baleia (mesomediterranean), Pinheiro da Cruz and Vila Nova de Milfontes (termomediterranean). All sampled acacia plants were located underneath a pine forest, with the exception of the latter case study, where we also sampled in a diversified habitat conditions associated with the margins of agriculture fields. According to the historical records, it was also possible to compare acacia samples from original plantations and recently invaded areas. We have used two molecular markers – inter-simple sequence repeats (ISSR) and microsatellites (SSR). ISSR analysis showed that A. longifolia populations at all sampled locations have similar levels of diversity and a relatively low differentiation (ΦPT=0.135). However, samples tended to cluster according to the regional (macro-scale) collection site. Microsatellites confirmed this low differentiation pattern. Our results indicate that differences in phenology and reproductive success observed during a previous study cannot be accounted for by genetic diversity and differentiation alone. Furthermore, our results taken together with historical information of A. longifolia suggest a single original introduction and a subsequent acclimation process.
- Article
(3311 KB) - Full-text XML
-
Supplement
(888 KB) - BibTeX
- EndNote
The genus Acacia comprises several species considered to be aggressive invaders, and the Australian species are considered important subjects of ecological invasion studies due to their introduction to several regions worldwide (e.g. Brazil, South Africa and Portugal), rendering them serious environmental threats on a global scale (Richardson and Rejmánek, 2011). Acacia longifolia (Andrews) Willd., also known as “Sydney Golden Wattle”, is a nitrogen fixing legume original from south-eastern Australia and Tasmania. This species is one of the most aggressive invaders worldwide and has been the subject of many studies which seek to understand its invasive mechanisms in order to prevent transfer to adjacent soils (e.g. Marchante et al., 2008; Richardson and Rejmánek, 2011; Ulm et al., 2017). It is known that the invasive capacity of A. longifolia is due to its ability to form a high number of root nodules in different soils and the establishment of a symbiotic relationship with the soil's mutualists (Rodríguez-Echeverría et al., 2008), including the genera Bradyrhizobium and Rhizobium (Rodríguez-Echeverría et al., 2011; Birnbaum et al., 2012), enabling spreading to nutrient-poor soils, such as the dune systems (Ulm et al., 2017). Also, A. longifolia promotes the success of its own offspring (Rascher et al., 2011) and has the ability to control available sunlight to other plants, eventually outcompeting them and occupying their space (Rascher et al., 2011). Furthermore, this species is capable of quick regeneration after fires because seed germination is actually promoted by the fire itself (Marchante et al., 2003). This is surely one of the reasons for its high invasiveness in Portugal, a country with a Mediterranean climate and frequent fires during the summer.
A. longifolia was introduced to the Portuguese coastlines by the forestry services at different periods between the late 19th and 20th century as a way to prevent the erosion of sand dunes (Marchante, 2011). Since its introduction, this species has invaded many adjacent environments including Pinus pinaster plantations, which has a high economic impact (Fernandes et al., 2015). A. longifolia's invasive success in Portugal is such that, in spite of its fairly recent introduction some 130 years ago, it can be found throughout the country where it is negatively impacting the diversity of native species (Marchante et al., 2003) and ecosystem functioning (Rascher et al., 2012; Ulm et al., 2017) such as changes in plant diversity (Holmes and Cowling, 1997; Lorenzo et al., 2012; Lazzaro et al., 2014), soil composition (Marchante et al., 2008; Souza-Alonso et al., 2015) and water availability (Lorenzo and Rodríguez-Echeverría, 2015).
Although there is a large distribution of this species under different conditions, its reproductive success is dependent on the site (Fernandes et al., 2015), in particular the flowering period, since it influences fruit and seed production (Pyšek and Richardson, 2008). A study on A. longifolia under two different environmental conditions – higher precipitation with lower average temperature (north of Portugal), and lower precipitation with higher average temperature (south of Portugal) – indicated large differences in the phenology and potential invasiveness impacts due to significant differences in flowering success and consequent seed production (Fernandes et al., 2015). Under typical Mediterranean climate and sparser forest canopy, acacias showed a shorter flowering period when compared to those under the more mesic Mediterranean conditions and denser forest canopy. This resulted in differences in reproductive success, with acacias under mesic conditions being more successful (Fernandes et al., 2015). These results suggest an influence of local environmental factors in the phenology of A. longifolia, particularly the temperature and forest management, which may cause anticipation in the phenological events (Fabio et al., 2010). Later, Morais and Freitas (2015) carried out a more extensive study, comparing the phenology of acacias from the north and south of Portugal and also from coastal (lower altitude) and inland acacias (higher altitude). The results once again show large differences in the phenology of A. longifolia in response to different environmental conditions, with acacias from the south of Portugal showing early phenological events, in particular the flowering period, due to the higher average temperature found at this location. These studies clearly show this species' ability to respond to new environmental conditions, and such local responsiveness associated with some degree of plasticity, might be one of the reasons for the invasion success of this species worldwide.
Despite all the studies mentioned above, there is limited information concerning A. longifolia on a molecular level, in particular the genetic variation and development of genetic structure after introduction. In recent years, the interest of molecular studies as a tool to elucidate genetic diversity for invasive species has increased. When a species is introduced to a new location, the environmental conditions it faces may be very different from the ones at its native location, and the introduced species has to overcome a number of challenges and adapt to the new environment (Schierenbeck and Ainouche, 2006). Furthermore, genetic diversity has great importance in the invasive capacity of a species, as was elegantly suggested by Lavergne and Molofsky (2007). However, despite its active role, genetic diversity might not be strictly necessary for invasion. Baker (1965) proposed that a species might be able to respond to different environmental conditions through phenotypic plasticity and little genetic evolution, coining the term “general-purpose genotype”, which has since then been the subject of many studies.
Molecular markers are an important and useful tool in molecular biology and can be used in studies of invasive species as means to identify source populations, determine invasive routes and elucidate invasive mechanisms and success (Handley et al., 2011). Many molecular markers have been used in population and phylogenetic studies in acacias, including inter-simple sequence repeats (ISSRs), simple sequence repeats (SSRs or microsatellites), chloroplastidial DNA and internal and external transcribed spacers (ITS and ETS respectively) (Thompson et al., 2012, 2015; Omondi et al., 2010, 2016; Harris et al., 2012; Miller et al., 2011; Le Roux et al., 2011, 2014; Fredua-Agyeman et al., 2008; Mulumba et al., 2012; Josiah et al., 2008; Ndlovu et al., 2013; Murphy et al., 2010; Brown et al., 2010, 2012). Furthermore, several SSRs markers have been developed for Acacia species, such as A. atkinsiana (Levy et al., 2014), A. brevispica (Otero-Arnaiz, 2005). A. dealbata (Guillemaud et al., 2015), Acacia (Senegalia) mellifera (Ruiz-Guajardo et al., 2007), A. nilotica (Wardill et al., 2004), A. mangium (Butcher et al., 2000), A. senegal (also known as Senegalia senegal) (Assoumane et al., 2009). However, to the best of our knowledge, SSR primers have not yet been described for A. longifolia.
In this study, we used two different types of molecular markers – ISSRs and SSRs – to assess the genetic diversity and differentiation of A. longifolia from three different locations in Portugal with similar sandy soil types but different climates and management conditions: Vila Nova de Milfontes and Pinheiro da Cruz (south), and Osso da Baleia (north). Furthermore, a detailed analysis was carried out within one of the three previous locations – Vila Nova de Milfontes – where three different sites were studied, including the site at which acacias were planted by the forestry services (original site). The aim of this study was to evaluate the genetic diversity and differentiation among this original site and other invaded locations to help summarize A. longifolia's invasive history in Portugal.
2.1 Plant material and characterization of study sites
Phyllodes (leaf-shaped petioles) from 25 samples of Acacia longifolia were collected in Osso da Baleia (OB) and Pinheiro da Cruz (PC), in the vicinity of a set of central geographic coordinates (Supplement: Tables S1 and S2). In Vila Nova de Milfontes, samples were collected in diversified habitat conditions (variation in a single location), including 14 samples from acacias that were originally planted (MFOld), 25 samples from the recently invaded sites (MFRec) and 14 samples scattered along this region (MFCL). A. saligna was included as an external control. For each individual, the geographic coordinates were determined (Table S3) and the geographical distribution was noted (Fig. 1).
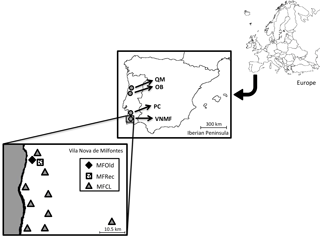
Figure 1Locations of sample collection. QM – Quiaios-Mira, OB – Osso da Baleia, PC – Pinheiro da Cruz, VNMF – Vila Nova de Milfontes, MFOld – site of plantation by the forestry services, MFRec – site adjacent to MFOld where acacias appeared more recently, MFCL – scattered sites of more recent acacias.
Field sites were established along the Atlantic coast under very different climatic conditions (variation along the Portuguese coast), corresponding to representative regions invaded with A. longifolia in the last decades. All studied sites were in dune ecosystems. Osso da Baleia has a mesomediterranean climate, with an average yearly precipitation of 841 mm and a typical pine forest (the major Pinus pinaster forest ecosystem in Portugal). The other two locations are south Lisbon, in a termomediterranean climate (average precipitation 400–600 mm) but differ in land-use: Pinheiro da Cruz, a forest type with a lower density typical of drier pine forest regions (Fernandes et al., 2015), and Vila Nova de Milfontes, an agroforestry system.
2.2 DNA extraction and quantification
DNA was extracted using a method developed by Doyle and Doyle (1987), modified by Weising et al. (1994) and adapted to A. longifolia as follows: phyllodes were macerated with liquid nitrogen using a mortar and a pestle (approx. half of a phyllode, 50–100 mg) and incubated at 65 ∘C with a 2 % CTAB extraction buffer. DNA was precipitated with pure ethanol at −20 ∘C and resuspended in TE buffer. After extraction, DNA was diluted to 10 µM and amplified through a PCR (polymerase chain reaction) in a BioRad T100 thermocycler.
2.3 Inter-simple sequence repeats (ISSRs)
Each PCR reaction took place in a total volume of 15 µL containing 6 ng of genomic DNA, 1 µM of each ISSR primer (STAB Vida, Portugal), 0.2 mM of dNTP mix (Promega, USA), 1 mg mL−1 of BSA (bovine serum albumin, Sigma, USA), 1.5 mM of MgCl2, 0.6 U of GoTaq Flexi DNA polymerase and 1× green GoTaq Flexi buffer (Promega, USA). A total of 31 primers were tested but only a few were selected for further analysis: 16 were selected for analysis of genetic diversity in the three collection sites and 11 were selected for a detailed study of this species' diversity in Vila Nova de Milfontes (Table S4). Primers that showed no amplification or low polymorphism level were excluded. Each PCR reaction included a negative control with all the components except the genomic DNA. As a routine procedure, each PCR included all accessions at the same time, using the same master mix. Reproducibility was confirmed by shared samples in both analyses, along the Portuguese coast and at single locations (MFOld and MFRec, Sect. 2.1), and by repeating amplification with two primers for all accessions. The PCR standard procedure consisted of an initial denaturation step at 95 ∘C for 5 min followed by 40 cycles of 45 s at 95 ∘C, 45 s at the corresponding annealing temperature for each primer (Table S4), and 1.5 min at 72 ∘C, followed by a final extension step at 72 ∘C for 7 min. For primers 840, 846, 901, 903 and 904, a touch-down programme varied in annealing temperatures, with 11 cycles at 57 ∘C, followed by 29 cycles at 52 ∘C (primers 901, 903 and 904), 53 ∘C for primer 846 or 54 ∘C for primer 840. The amplification products were separated by electrophoresis in a 2 % agarose gel with 2 µL of GreenSafe per 100 mL of agarose, using 1× TAE as both the gel buffer and the running buffer. Gels were run at 60 V for 4 h and were visualized in the GeneFlash UV transilluminator (Syngene, UK).
2.4 Simple sequence repeats (SSRs) or microsatellites
A total of 11 pairs of microsatellite primers designed for other Acacia species (Roberts et al., 2013; Forrest et al., 2015) were tested, and DCLOC and CPUH4 were selected for analysis of 97 accessions (Table S5). Each PCR reaction (15 µL) contained 10 ng of genomic DNA, 0.2 µM of labelled forward primer, 0.2 µM of reverse primer (STAB Vida), 0.2 mM of dNTP mix, 2 mM of MgCl2, 1 U of GoTaq Flexi DNA polymerase and 1× colourless GoTaq Flexi buffer (Promega, USA). Each PCR reaction included a negative control. The amplification consisted of an initial denaturation step at 95 ∘C for 5 min followed by 40 cycles of 30 s at 94 ∘C, 1 min at 59 ∘C, and 30 s at 72 ∘C, followed by a final extension step at 72 ∘C for 7 min. Twelve repeated samples were analysed for confirmation of amplification and reproducibility. These samples (and the negative controls) were loaded on 2 % agarose gel, run by electrophoresis at 80 V for 2 h and visualized as previously described. After confirmation of amplification, samples were analysed through capillary electrophoresis at a genotyping service provider (STAB Vida). For confirmation of the amplified motif, two samples were sequenced per locus.
2.5 Statistical analysis
Each ISSR gel image was analysed using GeneTools software (Syngene, UK) and only bands with sizes between 250 and 2400 bp were considered in the analysis. Reproducibility was confirmed by checking the resulting binary matrices between two independent ISSR amplifications. After obtaining the binary matrices, the data from all the primers were then combined and computed using NTSYS v2.1 (Rohlf, 2000) software to obtain the dendrograms by UPGMA (unweighted pair group method with arithmetic mean) with Dice's coefficient. Descriptive parameters (Nei's gene diversity, h, Nei, 1973, analogous to the expected heterozygosity under Hardy–Weinberg equilibrium; mean expected heterozygosity within subpopulations, HS; expected heterozygosity in total population, HT; and Wright's fixation index, FST) were computed using the AFLP-Surv v1.0 software (Vekemans et al., 2002) using a Bayesian method with a non-uniform prior distribution of allele frequencies and assuming a Hardy–Weinberg equilibrium (that is, FIS=0). Analysis of molecular variance (AMOVA) based on the ΦPT parameter and pairwise ΦPT matrixes were computed using GenAlEx v6.502 (Peakall and Smouse, 2006, 2012) software with 999 permutations. ΦPT is an analogue of FST.
SSR results were interpreted with Peak Scanner v1.0 software (Applied Biosystems, USA) with GS500(–250) as size standard. A Hardy–Weinberg equilibrium test, descriptive parameters (number of detected alleles, A; observed heterozygosity, Ho; expected heterozygosity, He; inbreeding coefficient, FIS; mean expected heterozygosity in total population over all loci, HT; and Wright's fixation index, FST) and allelic frequencies were computed using GenAlEx v6.502 as well as the AMOVA with 999 permutations based on the ΦPT parameter and the pairwise ΦPT matrix. The presence of null alleles was checked with Micro-checker v.2.2.3 software (Van Oosterhout et al., 2004).
3.1 Genetic diversity assessed through ISSR analysis
3.1.1 Variation along the Portuguese coast
A total of 31 ISSR primers were tested and 16 were selected to assess the genetic variability in Vila Nova de Milfontes (MFOld and MFRec), Osso da Baleia (OB) and Pinheiro da Cruz (PC). The choice of the primers was based on the amplification of clear and polymorphic bands, as shown in the agarose gel in Fig. S1 (Supplement). A total of 275 bands (243 excluding the external control) was obtained (Table S6), resulting in a dendrogram that shows a high genetic differentiation of all A. longifolia samples from the external control (EC) A. saligna (SDice=0.20) (Fig. 2). Separation among A. longifolia samples occurred at a much higher Dice's coefficient (first separation at SDice=0.63) and the clusters are all close together (last clustering at SDice=0.85), indicating that these samples shared a high degree of genetic similarity despite their geographical collection site. The dendrogram shows that samples clustered according to their geographical location, indicating some differentiation, but not all samples from the same location clustered together. The first separation on the dendrogram produces two clusters, I and II. Cluster I then subdivides into groups A and B, the former including all the samples from MFOld and three samples from MFRec (Fig. 2, circled in black). In group B two subgroups were considered, the first containing most of the samples from OB and subgroup 2 containing all samples from PC. Cluster II is divided into groups C and D, respectively containing most of the samples from MFRec and nine samples from OB.
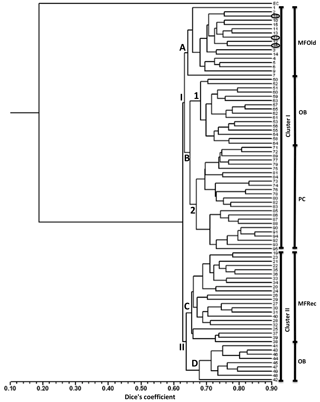
Figure 2Dendrogram obtained with ISSR markers with samples from Vila Nova de Milfontes (MFOld and MFRec), Osso da Baleia (OB) and Pinheiro da Cruz (PC). EC – external control, samples 1–15 – MFOld, samples 16–40 – MFRec, samples 41–65 – OB, samples 71–95 – PC. Samples circled in black are from MFRec but clustered together with samples from MFOld.
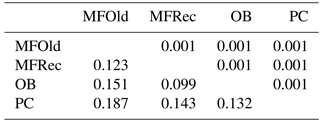
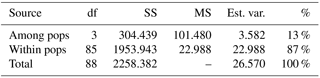
The computation of descriptive population genetic parameters allowed a better understanding of the genetic diversity throughout coastal Portugal (Table 1). The obtained mean Nei's gene diversity (h) is 0.190, with the highest value belonging to MFOld (h=0.213) and the lowest value belonging to PC (h=0.166), indicating that acacias found in PC were genetically less diverse than acacias found in MFOld. The computation of the FST showed a significantly low genetic differentiation, with a value of 0.070. AMOVA analysis (Table 2) based on the ΦPT value revealed that most variance occurred within populations (87 %) instead of among populations (13 %), and resulted in a significant ΦPT value of 0.135 (Table 1). Furthermore, to check the values of the ΦPT between each and every location, the pairwise ΦPT matrix was computed, with all obtained values being significant (Table 3). MFRec and OB were the most similar (ΦPT=0.099), while MFOld and PC were the most genetically different (ΦPT=0.187). Furthermore, MFOld and MFRec also presented some genetic differentiation (ΦPT=0.123), although they are geographically close (Fig. 1). Pairwise ΦPT values furthermore suggest that OB is more similar to MFRec (ΦPT=0.099) than to MFOld (ΦPT=0.151). PC presents the highest ΦPT values in general, indicating that this was the most differentiated location of all four.
Table 1Descriptive parameters of genetic variability of A. longifolia obtained with ISSR markers for samples from Vila Nova de Milfontes, Osso da Baleia and Pinheiro da Cruz. Samples from Vila Nova de Milfontes are shown in two subsets: MFOld and MFRec. No. PB – number of polymorphic bands without the external control; % PB – percentage of polymorphic bands. Standard error is shown in parenthesis.
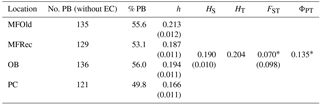
∗ , based on 500 permutations for FST and 999 for ΦPT.
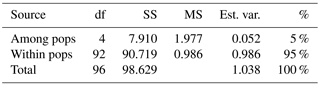
Table 2Analysis of molecular variance (AMOVA) of samples from Vila Nova de Milfontes (MFOld and MFRec), Osso da Baleia (OB) and Pinheiro da Cruz (PC) for ISSR variation. df – degrees of freedom, SS – sum of square, MS – mean square, est. var. – estimated variance, % – percentage of molecular variance. Pops is populations.
3.1.2 Variation in a single location
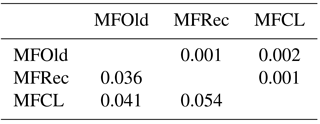
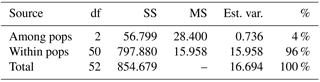
For the detailed analysis of the genetic variation in a single location, we used the material collected in Vila Nova de Milfontes, where 11 primers revealing high polymorphism were selected, generating a total of 137 bands, 128 excluding the external control (Table S7). The dendrogram presented in Fig. 3 again shows clear separation of the external control A. saligna from all the A. longifolia samples (SDice=0.31). The samples from Vila Nova de Milfontes shared roughly the same similarity level obtained previously (SDice=0.61–0.79 vs. SDice=0.63–0.85). However, the samples did not cluster according to the collection sites, MFOld, MFRec or MFCL. The mean Nei's gene diversity (h) is 0.261 and quite similar among sites, varying from h=0.275 (MFOld) to h=0.246 (MFRec). The global FST was 0.013 (Table 4). AMOVA analysis (Table 5) revealed that almost all variance occurred within (96 %) instead of among populations (4 %) and resulted in a significant ΦPT value of 0.044 (Table 4). The pairwise ΦPT matrix (Table 6) revealed that MFOld and MFRec were the most similar sites (ΦPT=0.036), while MFRec and MFCL were the most genetically different locations (ΦPT=0.054). All value obtained were significant.
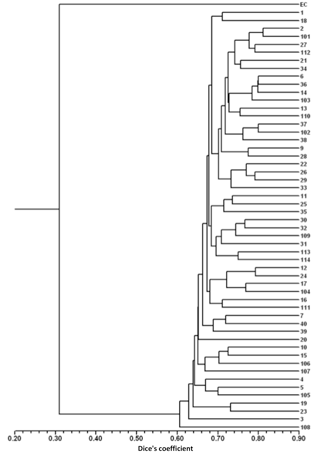
Figure 3Dendrogram obtained with ISSR markers of samples from Vila Nova de Milfontes (MFOld, MFRec and MFCL). EC – external control, samples 1–15 – MFOld, samples 16–40 – MFRec, samples 101–114 – MFCL.
Table 4Descriptive parameters of genetic variability of A. longifolia obtained with ISSR markers for samples from Vila Nova de Milfontes. No. PB – number of polymorphic bands without the external control; % PB – percentage of polymorphic bands. Standard error is shown in parenthesis.
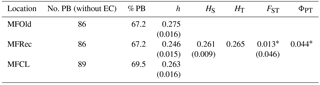
∗ , based on 500 permutations for FST and 999 for ΦPT.
Table 5Analysis of molecular variance (AMOVA) of samples from Vila Nova de Milfontes (MFOld, MFRec and MFCL) for ISSR variation. df – degrees of freedom, SS – sum of square, MS – mean square, est. var. – estimated variance, % - percentage of molecular variance. Pops is populations.
3.2 Genetic diversity assessed by microsatellite analysis
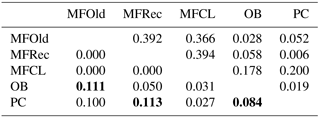
The initial screen of eleven pairs of primers allowed the selection of loci DCLOC and CPUH4 because they did not show unspecific amplifications and the amplified fragments had similar sizes to those previously described for other Acacia species. Locus DCLOC, described for A. loderi, has the repeat motif (CT)11 with an allele size range of 128–160 bp (Roberts et al., 2013). Locus CPUH4, described for A. melvillei, has the repeat motif (AT)13, with an allele size range of 112–115 bp (Forrest et al., 2015). The electropherograms obtained for locus DCLOC and CPUH4 resulted in a total of seven alleles in A. longifolia: alleles 98, 102 and 108 bp for DCLOC and alleles 93, 95, 99 and 121 bp for CPUH4. No private alleles were found within these loci. Locus DCLOC has two high-frequency alleles, 98 and 102, and a low-frequency allele, 108, which was absent from the PC location (Fig. 4). In locus CPUH4, two high-frequency alleles, 93 and 121 and two low-frequency alleles, 95 and 99, were identified. Allele 95 was absent from the MFRec and PC locations while allele 99 was absent from the MFCL site (descriptive statistics in Table 7). All five populations were on Hardy–Weinberg equilibrium, with the exception of MFCL, which failed the Hardy–Weinberg equilibrium test for locus CPUH4 (p value <0.01). The PC location is the one with the lowest He value (Table 7), reinforcing the idea that this population is less diverse than the remaining locations. The FIS values show that the OB and PC locations had an excess of heterozygotes, while all the other locations had slightly more homozygotes than heterozygotes. However, overall there is a slight excess of heterozygotes (average 0.039). There was no evidence of null alleles in any of the five locations with either locus analysed, according to the method that we used (Van Oosterhout et al., 2004). AMOVA analysis (Table 8) based on ΦPT revealed that very little variance is found among populations 5 % as opposed to within (95 %) populations. The FST value obtained for the SSR data (0.024, Table 7) proved to be statistically significant (), as did the ΦPT value (0.054, Table 7). The pairwise ΦPT matrix (Table 9), however, shows only three statistically significant values: between OB and MFOld, between PC and MFRec, and between PC and OB. The lowest pairwise ΦPT values were obtained among MFOld, MFRec and MFCL collection sites, and the PC location in general had high values of ΦPT compared to all other locations under study, the highest being with MFRec and statistically significant. Furthermore, the OB location has a lower ΦPT value with MFRec than with MFOld. In general, and as obtained with the ISSR data, the ΦPT values were low. The presented microsatellite data support the results obtained with the ISSR markers.
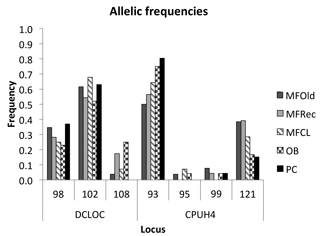
Figure 4Allelic frequencies of loci DCLOC and CPUH4 for samples from Vila Nova de Milfontes (MFOld, MFRec and MFCL), Osso da Baleia (OB) and Pinheiro da Cruz (PC).
Table 7Descriptive statistics for samples from Vila Nova de Milfontes (MFOld and MFRec), Osso da Baleia (OB) and Pinheiro da Cruz (PC) obtained with microsatellites. SE – standard error; HWE – p value for Hardy–Weinberg equilibrium test; ns – non-significant. No standard error computed for HT because of insufficient loci.
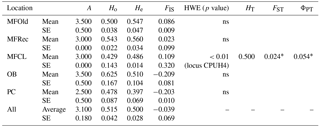
∗ , based on 999 permutations.
Table 8Analysis of molecular variance (AMOVA) of samples from Vila Nova de Milfontes (MFOld, MFRec and MFCL), Osso da Baleia (OB) and Pinheiro da Cruz (PC) for microsatellite loci variation. df – degrees of freedom, SS – sum of square, MS – mean square, est. var. – estimated variance, % – percentage of molecular variance. Pops is populations.
A. longifolia is an aggressive invasive species that, even in the relatively short period since its introduction to the Portuguese coastal region (6 to 7 decades ago), was able to spread throughout the coast and is now found virtually everywhere in Portugal, especially on the sand dunes in the Atlantic coastal areas. This study is, to the best of our knowledge, the first assessment of the genetic diversity of A. longifolia in an exotic region.
4.1 Genetic diversity and population differentiation
Analysis of the three main locations – OB, PC and VNMF – through ISSRs showed an overall low genetic diversity and a low, yet significant, genetic differentiation among the locations, resulting in a cluster analysis where samples grouped by sampling site (Fig. 2). The exception was OB, where samples were split between two different clusters, and the samples from the two locations MFOld and MFRec that, in spite of its geographic proximity (Fig. 1), clustered in two different parts of the dendrogram. However, three samples from MFRec still clustered with samples from MFOld (Fig. 2). Altogether, this low level of genetic differentiation and the pattern of clustering do not suggest the existence of several independent, well differentiated populations and, conversely, seems to indicate a single introduction of A. longifolia to Portugal that resulted from the plantation by the national forestry services. Since the plantation of this species occurred in several places along the Portuguese coastline at different times (from the late 19th century until the early 1970s, according to records), and based on our results, we now hypothesize that the forestry services might have used the same seed allotment or transferred seeds from one location to the others. Furthermore, our study suggests that this dispersion should have occurred relatively recently, as described by the historical records (Rei, 1925; Marchante, 2011), similarly to what has been reported with Vachellia (Acacia) karroo, a native woody species from South Africa that invades grasslands, which showed no genetic clustering and limited genetic differentiation (Taylor and Barker, 2012).
The genetic diversity observed in A. longifolia (h=0.166–0.213, Table 1) seemed to be within the range of values for other Acacia species living in their natural habitat, for example populations of A. senegal in Kenya (h=0.283, using ISSRs and RAPDs) (Josiah et al., 2008), A. caven (h=0.278, using AFLPs) and A. visco (h=0.200, using AFLPs) from Argentina (Pometti et al., 2012, 2016). However, it is important to emphasize that comparisons of genetic diversity among studies with different molecular markers have to be interpreted with care, since methods are sensitive to sample size and number of loci scored. Considering this, and without further analysis of genetic diversity of A. longifolia's native populations, our conclusions are necessarily provisional.
The statistical significance of FST and ΦPT for ISSRs can be the result of the relatively larger differentiation of the samples from the PC location and a by-product of a combination of founder effects and a strong drift in this location. Overall, the microsatellite results support the ones obtained with the ISSRs. The PC location shows the lowest He value and the highest significant pairwise FST values, showing that it is the most differentiated and least diverse of all locations studied. Also, OB has a statistically significant FST value when compared to MFOld, and once again the ISSR analysis has also shown a relatively high value between these two locations. Therefore, our ISSR analysis was robust.
Microsatellites have been used in studies of genetic variability of several Acacia species, either as native or invasive ranges. Studies in native A. senegal from Uganda (Mulumba et al., 2012) reported a mean expected heterozygosity similar to A. longifolia (He=0.479 and He=0.500 respectively). However, A. senegal had a higher and significant genetic differentiation among populations (FST=0.100), probably due to the fact that it is native. A. saligna and A. pycnantha native (Australian) populations revealed higher genetic differentiation (FST=0.330) compared to the invasive (South African) range (FST=0.132) (Thompson et al., 2012). Still, invasive A. saligna demonstrates greater differentiation than A. longifolia. For A. pycnantha, the values were similar for native and invaded species (FST=0.084 and FST=0.081 respectively) (Le Roux et al., 2013), and were of the same magnitude of those obtained in the present study.
4.2 Comparison between original plantation and invasion-dispersed populations
A. longifolia's ISSRs analysis was expanded in Vila Nova de Milfontes to a detailed study including all accessions (MFOld, MFRec and MFCL). The goal was to understand how the dispersal took place on a local scale and how (and if) that process influenced the genetic diversity and differentiation. No clustering was found based on the collection site (Fig. 3) and the values of the pairwise ΦPT matrix were low. The high genetic similarity of A. longifolia among Vila Nova de Milfontes samples is therefore explained by natural dispersal from the original MFOld stock to nearby locations either by natural (e.g. water, ants) or anthropogenic processes (e.g. agriculture, construction). These results also show no evidence of multiple introductions of A. longifolia to Vila Nova de Milfontes despite its widespread distribution in the study area (roughly 25 km; see Fig. 1), emphasizing this species' high invasive capacity.
4.3 Dispersal hypothesis and acclimation considerations
One of the most interesting results obtained in this study is that acacias from Osso da Baleia (OB) showed high similarity to samples from Vila Nova de Milfontes (VNMF) (Fig. 2), despite these two locations being the furthest apart in our study (Fig. 1). However, we found records of acacias' plantation in Quiaios-Mira sand dunes (QM in Fig. 1), a site located roughly 25 km north of OB (Rei, 1925; Marchante, 2011), which lead to the formulation of two hypotheses: (1) acacias spread from Quiaios-Mira to OB, in a similar situation to the one in VNMF mentioned above, and (2) the seeds used for plantation in Quiaios-Mira were also used for plantations by the forestry services in VNMF, which happened later (1924–1948 and 1960s–1970s respectively). These hypotheses need further molecular studies, with broader sampling for confirmation.
The most differentiated population studied was Pinheiro da Cruz (PC), probably due to a natural founder event based on a few individuals and subsequent genetic drift, consequently showing much less diversity than other man-made plantations from a larger stock of seeds. This population was relatively small in census size and under ecological constraints (e.g. drought) that seem to hamper its demographic and spatial expansion. It is also interesting to note that acacias under these conditions were less disturbed by anthropogenic activities. The combination of the founder event (probably by natural dispersal) and subsequent intense drift (due to small population size) explains both the small variability found in this population with both markers and the relatively high value of differentiation. However, this hypothesis needs to be checked in further studies with more information from other molecular markers and historical records.
The phenological results obtained by Fernandes et al. (2015) in A. longifolia from OB and PC locations showed a clear difference between the two sites, in particular in the flowering and reproductive successes. The question of whether this resulted from adaptation or acclimation to different climatic conditions was not clear. Our results do not support the hypothesis of large differentiation that could by itself be associated with the phenological differences already detected (Fernandes et al., 2015). However, we showed that acacias from the PC location presented the highest genetic differentiation and lowest genetic diversity compared to all other locations studied, which was supported through both molecular markers. It is certainly interesting to note that acacias from the PC location are also the acacias that showed lower reproductive success in the phenological study by Fernandes et al. (2015). Nevertheless, the small number of markers used and the random way they are scattered across the genome do not allow the establishment of a straight correlation between molecular diversity and phenological traits. Consequently, we did not expect that many of the markers were located in genomic regions associated with adaptive processes; as such they should essentially be neutral and mainly affected by the demographic processes. Another explanation for the phenological differences is that the ability of this alien species to respond to different environmental conditions is due to phenotypic plasticity, possibly fitting the general-purpose genotype concept proposed by Baker (1965) and further corroborated by Parker et al. (2003). Moreover, Baker (1965) also proposed the ideal weed description, which states that invasive species share traits, such as high reproductive success, high seed output and phenotypic plasticity. In fact, some traits are highly common among invasive species (Rejmánek and Richardson, 1996; Richardson and Pyšek, 2006), and A. longifolia does exhibit some of them. Therefore, genetic diversity does not seem to be a requirement for this species' invasive success. However, Baker's proposal propelled many studies on the subject and the conceptualization of several other hypotheses for invasive success (e.g. Rejmánek and Richardson, 1996; Zimmermann et al., 2014). Whether there is a causal relationship between diversity and reproductive success and whether other hypotheses other than the ideal weed description explain A. longifolia's invasive success, are certainly interesting and important topics for future research, as it might be relevant to determine the invasive process of this species and how to best decrease and eventually stop its invasion of the Portuguese coast.
In conclusion, A. longifolia along the Portuguese coast showed some degree of genetic differentiation among sampled locations. Moreover, there was genetic similarity when original planted acacias were compared with recent invader plants. Altogether, our results support the hypothesis that Portuguese acacia populations that are now spread along the coast all came from a single seed allotment origin, yet were undetermined, and a natural and/or subsequent man-made dispersal from the Portuguese coast to other areas. Finally, this study clearly suggests that the contrasting phenological patterns detected by other authors were not the result of independent colonization from different source native populations, nor were they a consequence of adaptation processes, demonstrating a wide capacity for phenotypic plasticity.
The code for each sample, the identification number and geographical coordinates for each location studied are available in the Supplement. We also provide, in the same file, detailed information about the ISSR and SSR primers used for this study, an example of a gel image obtained by amplification with an ISSR primer and tables with supplementary results of the ISSR analysis (e.g. percentage of polymorphic bands).
The supplement related to this article is available online at: https://doi.org/10.5194/we-18-91-2018-supplement.
The authors declare that they have no conflict of interest.
We thank Miguel Prado for his help with the samples from Vila Nova de
Milfontes and for the historical information provided, and Octávio S.
Paulo for his help with the analysis of microsatellite results. We are also
thankful for the financial support of CESAM (UID/AMB/50017 – POCI-01-0145-FEDER-007638),
FCT/MCTES through national funds (PIDDAC), the co-funding by the FEDER,
within the PT2020 Partnership Agreement and Compete 2020, and
UID/BIA/00329/2013.
Edited by: Adrian
Brennan
Reviewed by: two anonymous referees
Assoumane, A. A., Vaillant, A., Mayaki, A. Z., and Verhaegen, D.: Isolation and characterization of microsatellite markers for Acacia senegal (L.) Willd., a multipurpose arid and semi-arid tree, Mol. Ecol. Resour., 9, 1380–1383, https://doi.org/10.1111/j.1755-0998.2009.02669.x, 2009.
Baker, H. G.: Characteristics and modes of origin of weeds, in: The genetics of colonizing species, 1st edn., edited by: Baker, H. G. and Stebbins, G. L., Academic Press, New York, 147–168, 1965.
Birnbaum, C., Barrett, L. G., Thrall, P. H., and Leishman, M. R.: Mutualisms are not constraining cross-continental invasion success of Acacia species within Australia, Divers. Distrib., 18, 962–976, https://doi.org/10.1111/j.1472-4642.2012.00920.x, 2012.
Brown, G. K., Clowe, C., Murphy, D. J., and Ladiges, P. Y.: Phylogenetic analysis based on nuclear DNA and morphology defines a clade of eastern Australian species of Acacia ss. (section Juliflorae): the “Acacia longifolia group”, Aust. Syst. Bot., 23, 162–172, https://doi.org/10.1071/SB09037, 2010.
Brown, G., Murphy, D. J., Kidman, J., and Ladiges, P. Y.: Phylogenetic connections of phyllodinous species of Acacia outside Australia are explained by geological history and human-mediated dispersal, Aust. Syst. Bot., 25, 390–403, https://doi.org/10.1071/SB12027, 2012.
Butcher, P. A., Decroocq, S., Gray, Y., and Moran, G. F.: Development, inheritance and cross-species amplification of microsatellite markers from Acacia mangium, Theor. Appl. Genet., 101, 1282–1290, https://doi.org/10.1007/s001220051608, 2000.
Doyle, J. J. and Doyle, J. L.: A rapid DNA isolation procedure for small quantities of fresh leaf tissue, Phytochem. Bull., 19, 11–15, 1987.
Fabio, O., Carlo, S., Tommaso, B., Luigia, R., Bruno, R., and Marco, F.: Yield modelling in a Mediterranean species utilizing cause–effect relationships between temperature forcing and biological processes, Sci. Hortic., 123, 412–417, https://doi.org/10.1016/j.scienta.2009.09.015, 2010.
Fernandes, P., Antunes, C., Correia, O., and Máguas, C.: Do climatic and habitat conditions affect the reproductive success of an invasive tree species? An assessment of the phenology of Acacia longifolia in Portugal, Plant Ecol., 216, 343–355, https://doi.org/10.1007/s11258-014-0441-9, 2015.
Forrest, C. N., Roberts, D. G., Denham, A. J., and Ayre, D. J.: Microsatellite primers for vulnerable and thriving Acacia (Fabaceae) species from Australia's arid zone, Appl. Plant Sci., 3, 1400121, https://doi.org/10.3732/apps.1400121, 2015.
Fredua-Agyeman, R., Adamski, D., Liao, R. J., Morden, C., and Borthakur, D.: Development and characterization of microsatellite markers for analysis of population differentiation in the tree legume Acacia koa (Fabaceae: Mimosoideae) in the Hawaiian Islands, Genome, 51, 1001–1015, https://doi.org/10.1139/G08-087, 2008.
Guillemaud, T., Broadhurst, L., Legoff, I., Henery, M., Blin, A., Ducatillion, C., Ferrando, N., and Malausa, T.: Development of 23 polymorphic microsatellite loci in invasive silver wattle, Acacia dealbata (Fabaceae), Appl. Plant Sci., 3, 1500018, https://doi.org/10.3732/apps.1500018, 2015.
Handley, L.-J. L., Estoup, A., Evans, D. M., Thomas, C. E., Lombaert, E., Facon, B., Aebi, A., and Roy, H. E.: Ecological genetics of invasive alien species, Biol. Control, 56, 409–428, https://doi.org/10.1007/s10526-011-9386-2, 2011.
Harris, C. J., Dormontt, E. E., Le Roux, J. J., Lowe, A., and Leishman, M. R.: No consistent association between changes in genetic diversity and adaptive responses of Australian acacias in novel ranges, Evol. Ecol., 26, 1345–1360, https://doi.org/10.1007/s10682-012-9570-6, 2012.
Holmes, P. M. and Cowling, R. M.: Diversity, composition and guild structure relationships between soil-stored seed banks and mature vegetation in alien plant-invaded south African fynbos shrublands, Plant Ecol., 133, 107–122, https://doi.org/10.1023/A:1009734026612, 1997.
Josiah, C. C., George, D. O., Eleazar, O. M., and Nyamu, W. F.: Genetic diversity in Kenyan populations of Acacia senegal (L.) willd revealed by combined RAPD and ISSR markers, Afr. J. Biotechnol., 7, 2333–2340, 2008.
Lavergne, S. and Molofsky, J.: Increased genetic variation and evolutionary potential drive the success of an invasive grass, P. Natl. Acad. Sci. USA, 104, 3883–3888, https://doi.org/10.1073/pnas.0607324104, 2007.
Lazzaro, L., Giuliani, C., Fabiani, A., Agnelli, A. E., Pastorelli, R., Lagomarsino, A., Benesperi, R., Calamassi, R., and Foggi, B.: Soil and plant changing after invasion: the case of Acacia dealbata in a Mediterranean ecosystem, Sci. Total Environ., 497, 491–498, https://doi.org/10.1016/j.scitotenv.2014.08.014, 2014.
Le Roux, J. J., Brown, G. K., Byrne, M., Ndlovu, J., Richardson, D. M., Thompson, G. D., and Wilson, J. R.: Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) in South Africa, Divers. Distrib., 17, 861–871, https://doi.org/10.1111/j.1472-4642.2011.00784.x, 2011.
Le Roux, J. J., Richardson, D. M., Wilson, J. R., and Ndlovu, J.: Human usage in the native range may determine future genetic structure of an invasion: insights from Acacia pycnantha, BMC Ecol., 13, 37, https://doi.org/10.1186/1472-6785-13-37, 2013.
Le Roux, J. J., Strasberg, D., Rouget, M., Morden, C. W., Koordom, M., and Richardson, D. M.: Relatedness defies biogeography: the tale of two island endemics (Acacia heterophylla and A. koa), New Phytol., 204, 230–242, https://doi.org/10.1111/nph.12900, 2014.
Levy, E., Byrne, M., Coates, D. J., van Leeuwen, S. J., McArthur, S., Macdonald, B., and Gardner, M. G.: Isolation via 454 sequencing, and characterisation of microsatellite markers for the Pilbara endemic Acacia atkinsiana (Fabaceae), Conserv. Genet. Resour., 6, 585–587, https://doi.org/10.1007/s12686-014-0146-3, 2014.
Lorenzo, P. and Rodríguez-Echeverría, S.: Soil changes mediated by invasive Australian acacias, Ecosistemas, 24, 59–66, https://doi.org/10.7818/ECOS.2015.24-1.10, 2015.
Lorenzo, P., Pazos-Malvido, E., Rubido-Bará, M., Reigosa, M. J., and González, L.: Invasion by the leguminous tree Acacia dealbata (Mimosaceae) reduces the native understorey plant species in different communities, Aust. J. Bot., 60, 669–675, https://doi.org/10.1071/BT12036, 2012.
Marchante, H.: Invasion of portuguese dunes by Acacia longifolia?: present status and perspective for the future, PhD Thesis, University of Coimbra, Portugal, 2011.
Marchante, H., Marchante, E., and Freitas, H.: Invasion of the Portuguese dune ecosystems by the exotic species Acacia longifolia (Andrews) Willd.: Effects at the community level, in: Plant Invasion: Ecological Threats and Management Solutions, edited by: Child, L. E., Brock, J. H., Brundu, G., Prach, K., Pyšek, P., Wade, P. M., and Williamson, M., Backhuys Publishers, Leiden, 75–85, 2003.
Marchante, E., Kjøller, A., Struwe, S., and Freitas, H.: Short- and long-term impacts of Acacia longifolia invasion on the belowground processes of a Mediterranean coastal dune ecosystem, Appl. Soil Ecol., 40, 210–217, https://doi.org/10.1016/j.apsoil.2008.04.004, 2008.
Miller, J. T., Murphy, D. J., Brown, G. K., Richardson, D. M., and González-Orozco, C. E.: The evolution and phylogenetic placement of invasive Australian Acacia species, Divers. Distrib., 17, 848–860, https://doi.org/10.1111/j.1472-4642.2011.00780.x, 2011.
Morais, M. C. and Freitas, H.: Phenological dynamics of the invasive plant Acacia longifolia in Portugal, Weed. Res., 55, 555–564, https://doi.org/10.1111/wre.12177, 2015.
Mulumba, J., Nyakaana, S., and Jamnadass, R.: Genetic Diversity and Structure of Acacia senegal (L.) Willd. in Uganda, Int. J. Bot., 8, 107–116, https://doi.org/10.3923/ijb.2012.107.116, 2012.
Murphy, D. J., Brown, G. K., Miller, J. T., and Ladiges, P. Y.: Molecular phylogeny of Acacia s.s. (Mimosoideae: Leguminosae) – evidence for major clades and informal classification, Taxon, 59, 7–19, 2010.
Ndlovu, J., Richardson, D. M., Wilson, J. R., O'leary, M., and Le Roux, J. J.: Elucidating the native sources of an invasive tree species, Acacia pycnantha, reveals unexpected native range diversity and structure, Ann. Bot.-London, 111, 895–904, https://doi.org/10.1093/aob/mct057, 2013.
Nei, M.: Analysis of Gene Diversity in Subdivided Populations, P. Natl. Acad. Sci. USA, 70, 3321–3323, 1973.
Omondi, S. F., Kireger, E., Dangasuk, O. G., Chikama, B., Odee, D. W., Cavers, S., and Khasa, D. P.: Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya, Trop. Plant Biol., 3, 59–70, https://doi.org/10.1007/s12042-009-9037-2, 2010.
Omondi, S. F., Odee, D. W., Ongamo, G. O., Kanya, J. I., and Khasa, D. P.: Genetic consequences of anthropogenic disturbances and population fragmentation in Acacia senegal, Conserv. Genet., 17, 1235–1244, https://doi.org/10.1007/s10592-016-0854-1, 2016.
Otero-Arnaiz, A., Schnabel, A., Glenn, T. C., Schable, N. A., Hagen, C., and Ndong, L.: Isolation and characterization of microsatellite markers in the East African tree, Acacia brevispica (Fabaceae: Mimosoideae), Mol. Ecol. Notes, 5, 366–368, https://doi.org/10.1111/j.1471-8286.2005.00929.x, 2005.
Parker, I. M., Rodriguez, J., and Loik, M. E.: An evolutionary approach to understanding the biology of invasions: local adaptation and general-purpose genotypes in the weed Verbascum thapsus, Conserv. Biol., 17, 59–72, https://doi.org/10.1046/j.1523-1739.2003.02019.x, 2003.
Peakall, R. and Smouse, P. E.: genalex 6: genetic analysis in Excel. Population genetic software for teaching and research, Mol. Ecol. Notes, 6, 288–295, https://doi.org/10.1111/j.1471-8286.2005.01155.x, 2006.
Peakall, R. and Smouse, P. E.: GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update, Bioinformatics, 28, 2537–2539, https://doi.org/10.1093/bioinformatics/bts460, 2012.
Pometti, C. L., Bessega, C. F., Vilardi, J. C., and Saidman, B. O.: Landscape genetic structure of natural populations of Acacia caven in Argentina, Tree Genet. Genomes, 8, 911–924, https://doi.org/10.1007/s11295-012-0479-6, 2012.
Pometti, C. L., Bessega, C. F., Vilardi, J. C., Ewens, M., and Saidman, B. O.: Genetic variation in natural populations of Acacia visco, Plant Syst. Evol., 302, 901–910, https://doi.org/10.1007/s00606-016-1306-6, 2016.
Pyšek, P. and Richardson, D. M.: Traits Associated with Invasiveness in Alien Plants: Where Do we Stand?, in: Biological Invasions, edited by: Nentwig, D. W., Springer, Berlin, Heidelberg, 97–125, 2008.
Rascher, K. G., Große-Stoltenberg, A., Máguas, C., Meira-Neto, J. A. A., and Werner, C.: Acacia longifolia invasion impacts vegetation structure and regeneration dynamics in open dunes and pine forests, Biol. Invasions, 13, 1099–1113, https://doi.org/10.1007/s10530-011-9949-2, 2011.
Rascher, K. G., Hellmann, C., Máguas, C., and Werner, C.: Community scale 15N isoscapes: tracing the spatial impact of an exotic N2-fixing invader, Ecol. Lett., 15, 484–491, https://doi.org/10.1111/j.1461-0248.2012.01761.x, 2012.
Rei, M. A.: Arborização da Serra da Boa Viagem da Figueira da Foz (Subsídios para a sua história): 1911–1924, Tipografia Popular, Figueira da Foz, 1925.
Rejmánek, M. and Richardson, D. M.: What Attributes Make Some Plant Species More Invasive?, Ecology, 77, 1655–1661, https://doi.org/10.2307/2265768, 1996.
Richardson, D. M. and Pyšek, P.: Plant invasions: merging the concepts of species invasiveness and community invisibility, Prog. Phys. Geog., 30, 409–431, https://doi.org/10.1191/0309133306pp490pr, 2006.
Richardson, D. M. and Rejmánek, M.: Trees and shrubs as invasive alien species – a global review, Divers. Distrib., 17, 788–809, https://doi.org/10.1111/j.1472-4642.2011.00782.x, 2011.
Roberts, D. G., Forrest, C. N., Denham, A. J., and Ayre, D. J.: Microsatellite markers for vulnerable Australian aridzone Acacias, Conserv. Genet. Resour., 5, 199–201, https://doi.org/10.1007/s12686-012-9767-6, 2013.
Rodríguez-Echeverría, S., Crisóstomo, J. A., Nabais, C., and Freitas, H.: Belowground mutualists and the invasive ability of Acacia longifolia in coastal dunes of Portugal, Biol. Invasions, 11, 651–661, https://doi.org/10.1007/s10530-008-9280-8, 2008.
Rodríguez-Echeverría, S., Le Roux, J. J., Crisóstomo, J. A., and Ndlovu, J.: Jack-of-all-trades and master of many? How does associated rhizobial diversity influence the colonization success of Australian Acacia species?, Divers. Distrib., 17, 946–957, https://doi.org/10.1111/j.1472-4642.2011.00787.x, 2011.
Rohlf, F. J.: NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.1, Exeter Software, Setauket, New York, 2000.
Ruiz-Guajardo, J. C., Otero-Arnaiz, A., Taylor, T., Stone, G., Glenn, T. C., Schable, N. A., Miller, J. T., Preuss, S., and Schnabel, A.: Isolation of polymorphic microsatellite markers in the sub-Saharan tree, Acacia (Senegalia) mellifera (Fabaceae: Mimosoideae), Mol. Ecol. Notes, 7, 1138–1140, https://doi.org/10.1111/j.1471-8286.2007.01809.x, 2007.
Schierenbeck, K. A. and Ainouche, M. L.: The role of evolutionary genetics in studies of plant invasions, in: Conceptual Ecology and Invasion Biology: Reciprocal Approaches to Nature, edited by: Cadotte, M. W., Mcmahon, S. M., and Fukami, T., Kluwer Academic Publishers, Dordrecht, 193–221, 2006.
Souza-Alonso, P., Guisande-Collazo, A., and González, L.: Gradualism in Acacia dealbata Link invasion: impact on soil chemistry and microbial community over a chronological sequence, Soil Biol. Biochem., 80, 315–323, https://doi.org/10.1016/j.soilbio.2014.10.022, 2015.
Taylor, C. L. and Barker, N. P.: Species limits in Vachellia (Acacia) karroo (Mimosoideae: Leguminoseae): Evidence from automated ISSR DNA “fingerprinting”, S. Afr. J. Bot., 83, 36–43, https://doi.org/10.1016/j.sajb.2012.07.014, 2012.
Thompson, G. D., Bellstedt, D. U., Byrne, M., Millar, M. A., Richardson, D. M., Wilson, J. R., and Le Roux, J. J.: Cultivation shapes genetic novelty in a globally important invader, Mol. Ecol., 21, 3187–3199, https://doi.org/10.1111/j.1365-294X.2012.05601.x, 2012.
Thompson, G. D., Bellstedt, D. U., Richardson, D. M., Wilson, J. R., and Le Roux, J. J.: A tree well travelled: global genetic structure of the invasive tree Acacia saligna, J. Biogeogr., 42, 305–314, https://doi.org/10.1111/jbi.12436, 2015.
Ulm, F., Hellmann, C., Cruz, C., and Máguas, C.: N∕P imbalance as a key driver for the invasion of oligotrophic dune systems by a woody legume, Oikos, 126, 231–240, https://doi.org/10.1111/oik.03810, 2017.
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P., and Shipley, P.: MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data, Mol. Ecol. Notes, 4, 535–538, https://doi.org/10.1111/j.1471-8286.2004.00684.x, 2004.
Vekemans, X., Beauwens, T., Lemaire, M., and Roldán-Ruiz, I.: Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size, Mol. Ecol., 11, 139–151, https://doi.org/10.1046/j.0962-1083.2001.01415.x, 2002.
Wardill, T. J., Scott, K. D., Graham, G. C., and Zalucki, M. P.: Isolation and characterization of microsatellite loci from Acacia nilotica ssp. Indica (Mimosaceae), Mol. Ecol. Notes, 4, 361–363, https://doi.org/10.1111/j.1471-8286.2004.00697.x, 2004.
Weising, K., Nybom, H., Pfenninger, M., Wolff, K., and Meyer, W.: DNA Fingerprinting in Plants and Fungi, 1st edn., CRC Press, Boca Raton, 1994.
Zimmermann, H., Brandt, P., Fischer, J., Welk, E., and von Wehrden, H.: The Human Release Hypothesis for biological invasions: human activity as a determinant of the abundance of invasive plant species [version 2; referees: 2 approved], F1000Research, 3, 109, https://doi.org/10.12688/f1000research.3740.2, 2014.