the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Do mycorrhizal fungi create below-ground links between native plants and Acacia longifolia? A case study in a coastal maritime pine forest in Portugal
Pedro Carvalho
Rui Martins
António Portugal
M. Teresa Gonçalves
Maritime pine forests are a major ecosystem throughout the Portuguese coast and are severely affected by the invasion of Acacia longifolia. The presented study investigated the diversity of ectomycorrhizal fungi (ECM) of major plant species in these ecosystems to find possible links between Pinus pinaster, three native Cistaceae shrubs and the Acacia invasive species. We successfully identified 13 ECM fungal taxa. Within those, two species from the order Helotiales were found in all plant species, and over half of the fungal ECM species found in Pinus pinaster were also common to the Cistaceae shrubs. Network analysis points to the Cistaceae shrubs having a central role in these below-ground communities, therefore enforcing the idea that they are key to these communities and should not be underestimated. Our results also point to the evolving role of invasive plant species in the ecosystem dynamics in the rhizosphere, which host fungal species that are common to native plants, although it is not yet clear whether these fungal taxa are native or a consequence of the presence of Acacia longifolia.
- Article
(1067 KB) - Full-text XML
-
Supplement
(404 KB) - BibTeX
- EndNote
Plants can obtain many benefits from establishing mycorrhizal associations, for instance, better access to nutrient and water supplies and higher resilience to biotic and abiotic stresses (Smith and Read, 2006). Although there are some exceptions, the majority of ectomycorrhizal (ECM) fungal species are not truly host-specific and can colonize the roots of several individuals from different plant species, from trees to shrubs and even herbaceous plants (Bruns et al., 2002; Buscardo et al., 2012; Dickie et al., 2004; Ishida et al., 2007; Kennedy et al., 2003). The fundamental role of ECM fungi in ecosystem dynamics, nutrient cycling and plant performance is well established (Itoo and Reshi, 2014; Smith and Read, 2006) and approximately 25 000 species of fungi have been described as being able to establish ectomycorrhizal associations (Rincón et al., 2015; Tedersoo et al., 2010; Tedersoo and Smith, 2013),
Pinus pinaster, the “maritime pine”, is distributed along the western Mediterranean basin (Campelo et al., 2015), inhabiting mainly acid and silicon coastal soils (Berthier, 2001). It is one of the most important forest species in Portugal (Campelo et al., 2015; Monteiro-Henriques et al., 2016), covering 27.3 % of the forested area and being the most extensively distributed forest species (Godinho-Ferreira et al., 2005). All Pinaceae species are reported as establishing mainly ectomycorrhizas, although some studies describe the occurrence of arbuscular mycorrhizas (AM) in Pinus spp. and in other Pinaceae (e.g. Horton et al., 1998). In recent years the ectomycorrhizal fungal communities of Pinus spp. have been studied through molecular approaches using individual ectomycorrhizal root tips, soil and whole roots (Buscardo et al., 2010, 2011, 2012; Cox et al., 2010; Jarvis et al., 2013; Pestaña Nieto and Santolamazza Carbone, 2009; Rincón et al., 2015; Walbert et al., 2010). Maritime pine forests are a major ecosystem throughout the Portuguese coast and are severely affected by the invasion of Acacia longifolia.
The south-western Australian species Acacia longifolia is one of the most prolific invasive species in Portugal (Marchante et al., 2003). Acacia species are known to form AM and ECM associations, with AM being predominantly found in both native and invaded areas (Aswathappa et al., 1987; Rodríguez-Echeverría et al., 2009). To the extent of our knowledge, there are no reports of ECM associations with this species in Portugal.
Cistaceae shrubs are common understory species in the Mediterranean basin. They can establish both ECM and AM (Smith and Read, 2006) and have been reported to share ECM fungi with co-occurring Pinus pinaster (Buscardo et al., 2012). From the 12 species of the Mediterranean genus Cistus occurring in the Iberian Peninsula (Águeda et al., 2006; Alonso Ponce et al., 2011), nine are native in Portugal and compose 7 % of the forested area (Godinho-Ferreira et al., 2005). Being woody, evergreen and pyrophytic shrubs (Arrington and Kubitzki, 2003; Comandini et al., 2006) all species of Cistus are among the first colonizers after a disturbance event (e.g. fire or grazing) and thus are pioneer species of ecological successions (Águeda et al., 2006; Alonso Ponce et al., 2011; Comandini et al., 2006). About 230 species of Ascomycota and Basidiomycota fungi have been described as ECM symbionts of Cistus sp, with 35 being Cistus-specific mycobionts nearly all belonging to Russulaceae and Cortinariaceae families (Comandini et al., 2006). Halimium halimifolium is another Mediterranean Cistaceae shrub that occurs in sandy soil, becoming the dominant species in sand ridges where the water table depth ranges from 2 to 4 m (Zunzunegui et al., 2002). Based on a fruiting body survey, 12 ECM fungal species have been identified associated with H. halimifolium in Corsica (Taudiere et al., 2015), of which four species were also associated with Pinus pinaster.
We hypothesized that there is a high degree of similarity between the ECM fungal communities associated with the Cistaceae species and the ones associated with the maritime pine. Furthermore, we looked for putative ECM symbionts in the root system of nearby Acacia longifolia individuals with the aim of evaluating whether it shares putative symbionts with Cistaceae shrubs and with the dominant tree, Pinus pinaster.
In this study we investigated the ECM fungal communities of the most frequent plant hosts in a coastal maritime pine forest, namely Pinus pinaster, three Cistaceae shrubs (Cistus salviifolius, Cistus psilosepalus and Halimium halimifolium), and also the invasive species (Acacia longifolia). We achieve this by sorting ectomycorrhizal root tips into “ad hoc morphological groups” and carrying out further molecular identification by DNA barcoding using ITS sequencing (Schoch et al., 2012; Seifert, 2009). We observed a high degree of similarity between the ECM communities found in Pinus pinaster and in the Cistaceae shrubs. Although Acacia longifolia had a less diverse ECM fungal community, the few detected common ECM fungal species imply that it is already part of the below-ground dynamics of the fungal communities.
2.1 Study site
The study area was an even-aged managed forest of Pinus pinaster located in the central coast of Portugal (40.35834∘ N, 8.81903∘ W). The area has a typical Mediterranean climate with oceanic influence and a marked summer drought. The average annual temperature and precipitation are 16.2 ∘C and 953 mm and the soil is acidic with a sandy texture and low water-holding capacity from the order Inceptisol (Campelo et al., 2015). The area is dominated by Pinus pinaster trees with an average age of 45 years and a high occurrence of the exotic invasive Acacia longifolia. Cistus psilosepalus, Cistus salviifolius, Halimium halimifolium and Corema album are the most common understory shrubs.
2.2 Root sampling and analysis
Three individuals of each species, at distances of at least 1 m from each other, were randomly marked, and root samples were collected in mid-November 2015 using the methodology described by Pestaña Nieto and Santolamazza Carbone (2009). Briefly, samples were collected by digging around the tree or shrub, in all four cardinal points, using a trowel and following the root system from the base of each plant to a maximum depth of 10–20 cm for a period of about 1 h per tree. For each tree, the samples collected from the four cardinal points were mixed in a composite sample. Roots were collected by selecting those with a higher number of fine secondary roots surrounded by soil clods, indicative of a higher number of ectomycorrhizal root tips, and placed carefully along with soil clods into plastic bags for further laboratory analysis. Soil was separated from samples using a sieve, roots were weighed and subsequently washed and stored at 4 ∘C. To standardize the sampling effort, a total of 200 mg of fresh root weight was surveyed for each sample. Root samples of each individual were analysed under a stereo microscope and ECM root tips sorted into ad hoc morphological groups (Dickie and Reich, 2005) based on shape, ramifications, texture, colour and the presence of emanating hyphae or rhizomorphs (Agerer, 2001) (Table 1). Photographs of the morphotypes were obtained using Leica© EZ4 HD stereo microscope and Leica© LAS EZ software (Figure S1 in the Supplement).
2.3 Molecular identification
DNA extraction was performed for each morphotype, 1 mm2 of tissue was used for REDExtract-N-Amp™ (SIGMA-ALDRICH© Company) modified protocol, 20 µL of Extract solution was added to ectomycorrhizal tips and they were incubated to 94 ∘C for 10 min followed by 13 min at 60 ∘C and 15 min at 10 ∘C, 20 µL of dilution solution was added afterwards. PCR was performed using the primer pair ITS1F/ITS4 (Gardes and Bruns, 1993; White et al., 1990) and JumpStart™ Taq DNA polymerase without MgCl2, which was added subsequently according to manufacturer's instructions(SIGMA-ALDRICH© Company). 1 µL of the extraction was used as DNA template. PCR parameters were the following: 1 step of 94 ∘C for 5 min, 33 cycles of 94 ∘C for 45 s, 57 ∘C for 35 s and 72 ∘C for 35 s and 1 step of 72 ∘C for 10 min.
Amplifications were checked by electrophoresis in 2 % agarose gel. Single-banded samples were sequenced using the ABI PRISM© BigDye™ terminator cycle sequence reaction kit (Applied Biosystems©, Perkin Elmer).
Table 2Results from the BLAST analysis based on ITS sequences obtained from field-collected ECM root tips. The most similar sequences from NCBI are represented by their accession numbers. Species identification was done at sequence similarity higher than 97.
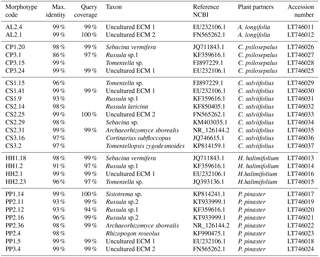
The obtained sequences were edited using Geneious® software, sequences quality (HQ) was assessed and low-quality sequences were discarded. Basic Local Alignment Search Tool (BLAST) was performed with the National Center for Biotechnology Information (NCBI) database to confirm species taxonomic identification (Altschul et al., 1997). Species identification was achieved using 97 % sequence similarity. Identical sequences, i.e. with sequence similarity higher than 99 %, were grouped and only one representative of each group was considered in subsequent analyses. Sequences obtained in this study were submitted to the European Nucleotide Archive (ENA) (Table 2). Sequences were aligned using the MUSCLE algorithm available in MEGA7® software. Phylogenetic analysis of the obtained sequences, together with sequences retrieved from online databases (GenBank), was performed using maximum likelihood methods with the GTR model of evolution, which was selected as the best-fitting model using the Akaike information criterion (AIC) in jModelTest (Guindon and Gascuel, 2003; Posada, 2008). The phylogenetic analysis was performed using PhyML (Guindon and Gascuel, 2003) in Phylemon 2.0 (Sánchez et al., 2011). Branch support was assessed using the bootstrap likelihood ratio test with 1000 repetitions. The resulting tree was represented using the R software (R Development Core Team, 2011) package ggtree from BioConductor (Yu et al., 2017) (Fig. 1).
2.4 Data analysis
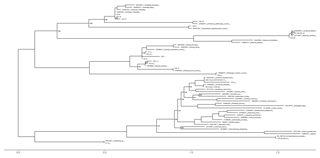
Figure 1Maximum likelihood tree based on ITS sequences obtained from field-collected ECM root tips and reference sequences obtained from Genbank (included with their accession numbers). Sequences obtained in this study are named in Table 1. Sequences with similarity > 99 % are grouped and listed together. Numbers at the nodes are values for branch support estimated using the bootstrap likelihood ratio test (1000 resamplings, values 0–1, values above 0.98 are not displayed for easier viewing). The scale bar indicates the evolutionary distances.
To characterize and compare the fungal communities associated with each plant species we calculated the Shannon index (H′) of diversity, using abundance as the number of ECM tips in each identified morphotype group, according to , where pi is the proportion of each fungal species in the community (Shannon and Weaver, 1949). Based on the results, the Pielou index (E) of evenness was calculated according to , where S is defined as the total number of species in the community (Species richness). For this, values closer to the unit indicate a community where the ECM tips are equally distributed among the species identified. The Margalef index (Dm) for species richness was calculated as follows: , where N is defined as the total number of ECM tips identified (Gamito, 2010) (Table 3). All indexes were obtained using R software (R Development Core Team, 2011) with the package vegan (Dixon, 2003).
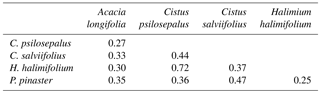
To analyse community composition and similarity, we resorted to the Bray–Curtis index (BC) for all pairwise combinations of plant species (Chao et al., 2005) (Table 4). This index estimates the similarity between communities. With these results we obtained a cladogram (Fig. 2), which allows the results to be clearly viewed using the R package vegan. Community composition comparisons can be seen in the ordination analysis, performed using a non-metric multidimensional scaling (NMDS). The two dimensions of separation of the communities are represented in graphic form (Fig. 3) with stress values of 0.033, using the R software package vegan.
Table 3Ecological indexes for each plant species considering the identified ECM symbionts. S is the total number of species, N is the total number of ECM tips, H′ is the Shannon–Wienner index, E is the Pielou index and D is the Margalef index.
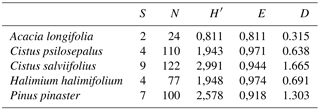
Table 4Similarity matrix between the ECM community associated with each plant species studied and calculated using the Bray–Curtis index of dissimilarity.
To further understand the relationship between the communities associated with each species, we computed the Euclidean distance between each fungal plant community, obtaining a metric distance matrix that was used to create a weighted and undirected network, using the R package qgraph (Epskamp et al., 2012) (Fig. 4). This algorithm allows the Euclidean distances to be visualized by resizing and shading the edges (Euclidean distances) between each node (plant-associated community) according to the values of each edge that translate the number of commonly identified species as well as the relationships between identified species in each community.
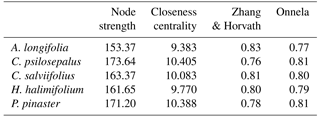
In order to further understand the network, we obtained values for centrality, specifically the value for betweenness centrality (node strength). This gives us a measure for centrality, not only a topological one but also one that influences the network of each node. The shortest path of the edges through the node are taken into account, which is found by the minimal sum of the edges that link any pairs of nodes in the network, as well as the weight of the edges, given by the Euclidean distances. If a high number of shortest paths that pass through a node it is considered a central node in the network. We also obtained the value for closeness centrality, which is calculated based on the sum of the lengths of the shortest paths between the node and all the other nodes in the graph, so that a central node is closer to all the other nodes. This is computed by the R package qgraph (Table 5). Moreover, two clustering coefficients were determined for each community: the Zhang and Horvath's weighted clustering coefficient (Zhang and Horvath, 2005) and Onnela's clustering coefficient (Onnela et al., 2005).
Table 5Topological indexes based on the network obtained with the Euclidian distances between each plant fungal community. Betweenness centrality, represented here by the node strength, closeness centrality and clustering coefficients (Zhang and Horvath; Onnela) indicate the importance and influence of each community in the network.
From 433 observed root tips, the morphological sorting resulted in 27 morphotypes for all plant species. After PCR sequencing and BLAST a total of 13 fungal taxa were identified (Table 2): two Ascomycota in association with Acacia longifolia, four Basidiomycota and three Ascomycota in association with Pinus pinaster, three Basidiomycota and one Ascomycota in association with Cistus psilosepalus, six Basidiomycota and three Ascomycota in association with Cistus salviifolius, and three Basidiomycota and one Ascomycota in association with Halimium halimifolium (Table 2).
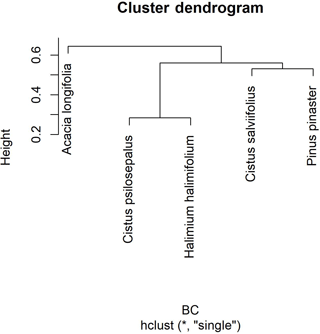
Figure 2Cluster dendrogram using average linkage between groups, given by the Bray–Curtis dissimilarity index matrix. The y axis shows rescaled distance cluster combinations.
Identification up to species level was not possible for 19 morphotypes that corresponded to four taxa (Table 2). Morphotypes identified as “Uncultured ECM 1” and “Uncultured ECM 2” belonged to the Helotiales order (Fig. 2). Two unidentified Russula species were named Russula sp. 1 and Russula sp. 2 and were closely related to Russula sardonia and Russula cascadensis, respectively (Fig. 1).
The most frequent genus was Russula, with three different identified taxa, and the most frequent taxon was the Uncultured ECM 1, found in all plant-associated communities.
Species richness and diversity, calculated according to the Margalef index and Shannon index, were the highest for Cistus salviifolius and Pinus pinaster while the lowest values were found in Acacia longifolia, Halimium halimifolium and Cistus psilosepalus communities show to be the most even.
The highest values of similarity were found between Halimium halimifolium and Cistus psilosepalus, which was an expected result since they host the same fungal species. The lowest levels were found between Pinus pinaster and Halimium halimifolium. When plotted, similar clusters were shown between Halimium halimifolium and Cistus psilosepalus, and between Cistus salviifolius and Pinus pinaster, with Acacia longifolia hosting the most dissimilar community (Table 3; Fig. 2). NMDS analysis showed that the plant-related communities were separated in two dimensions. The observed separations support the Bray–Curtis cladogram, showing the cluster of Halimium halimifolium with Cistus psilosepalus and Pinus pinaster with Cistus salviifolius as well as the separation of Acacia longifolia (Figs. 2 and 3).
Pinus pinaster shared four species with Cistus salviifolius (Uncultured ECM1, Uncultured ECM2, Archaeozhizomyces borealis and Russula sp. 1), two with Cistus psilosepalus (Uncultured ECM1 and Russula sp. 1) and two with Halimium halimifolium (Uncultured ECM 1 and Russula sp. 1). Over half of the fungal species found in Pinus pinaster were also present in the Cistaceae shrubs. The fungal species found in Acacia longifolia, Uncultured ECM 1 and Uncultured ECM 2, were also found in Cistaceae shrubs and in Pinus pinaster.
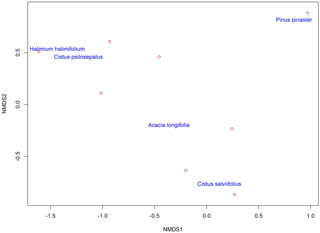
Figure 3Non-metric multidimensional scaling (NMDS) of the plant-associated fungal communities, showing the clustering of similar communities in the two dimensions represented here. Plant communities are represented by the name of the plant species. Each red circle represents one identified fungal taxa except for Pinus pinaster, where the red circle represents three different fungal taxa, and Cistus salviifolius, where the red circle represents four fungal taxa.
The network analysis suggests that Cistus psilosepalus is the most central and important node in the network, as shown by the topological indexes. On the other hand, Acacia longifolia is the least central and weakest node, but it showed the highest value in one of the clustering indexes.
We identified 13 different fungal taxa associated with the most common ECM host plants in the studied habitat. ECM fungal species associated with Halimium halimifolium are described for the first time and the results are in line with those obtained with above-ground fruiting body surveys (Taudiere et al., 2015).
By taking into account the small scale of this study and comparing our results with those obtained in a similar habitat (Pestaña Nieto and Santolamazza Carbone, 2009), we may conclude that this ecosystem has an appreciable diversity of ECM fungi and that most of the identified fungal species were common to two or more plant partners. These results are in agreement with previous studies showing the existence of common ECM fungi partners of Pinus pinaster and Cistaceae shrubs, suggesting the existence of a common mycorrhizal network (CMN) in this ecosystem in which putative partners have been identified.
Our study is also among the first to report ECM fungi associated with Acacia longifolia outside its native range. The above-ground negative impact of this invasive species in Portuguese forests is well documented (e.g. Marchante et al., 2015). Some authors have already investigated the below-ground symbiotic relations of A. longifolia (Rodríguez-Echeverría, 2010; Rodríguez-Echeverría et al., 2009). They were able to correlate its invasive success to the introduction of exotic rhizobia and to the ability to profusely nodulate with native or invasive bacteria, giving a glimpse into the effects and the ecology of invasive species in the new range.
In the present study, in contrast with the data available in terms of bacteria and AM fungi, we investigated another symbiotic association of A. longifolia, and our results pointed out another facet of the invasiveness of this species. We hypothesized that Acacia longifolia could already be a member of the community as the host of a fungal species that could colonize neighbouring native plant species. Our results support this hypothesis by showing that two fungal taxa were found both in Acacia longifolia and in the native species. This observation places Acacia longifolia within the network formed by the fungal species, although it is an outlink in the broader studied plant community as shown in the cladogram (Fig. 2). The ordination test also showed a clear separation from the other communities in both dimensions (Fig. 3).
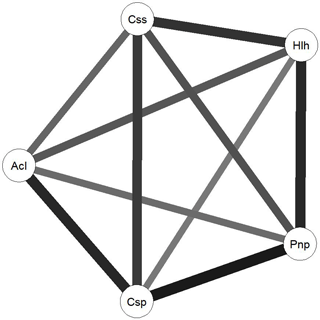
Figure 4Network diagram created using the R package qgraph. Circles indicate plant-related communities (nodes) and grey lines (edges) link plant-related communities based on the Euclidean distances between them. Weight of each line represents the weight of each edge in terms of the Euclidean distance results between each node. Css is Cistus salviifolius, Hlh is Halimium halimifolium, Pnp is Pinus pinaster, Csp is Cistus psilosepalus and Acl is Acacia longiolia.
The lack of information regarding the identity of some fungal taxa, particularly the Uncultured ECM 1 and Uncultured ECM 2, undermined further conclusions on the putative role of the invasion in these communities (Vrålstad et al., 2002), Moreover, the lack of information about the fungal associations of A. longifolia in its native range and in other older places of introduction, leave open the hypothesis that Acacia longifolia may be acting as a vector of dispersion and possibility of an invasion of fungal taxa to and from the invaded locations, as has been observed for bacterial symbionts (Rodríguez-Echeverría, 2010). The hosts of native fungal species may be expanded by establishing themselves as new hosts, deeply influencing the dynamics of these species (Pringle et al., 2009).
More studies should be conducted to further characterize the fungal communities of Acacia longifolia, both in their native location and in the places where they have been introduced, with varying levels of invasiveness.
In this study we described a novel host of the recently identified fungus Archaeorhizomyces borealis (Menkis et al., 2014), Cistus salviifolius, which is also its first non-gymnosperm host. It is possible that the structural similarities of the cortical Hartig net described for both the genus Cistus and gymnosperms hosts (Smith and Read, 2006) may explain this novel combination.
The high number of ECM fungal species shared between the studied plant species suggests that the role of the Cistaceae shrubs in these ecosystems is important. Moreover, the high similarity between the ECM fungal species found in the three surveyed species may indicate a certain specificity at the family level, since they harbour fungal species that also colonize pines. These shrubs can be of crucial importance as a reservoir for ECM fungal species after disturbances that affect mainly tree species, such as logging or recurrent wildfires (Buscardo et al., 2012). Contrary to what we expected, the dominant tree species, Pinus pinaster, is neither the most central nor the strongest node of the network, which is mainly driven by the high number of species shared between the Cistaceae shrubs and their weight on the network. Cistus psilosepalus is the most central node, mainly because all the associated species are the same for Halimium halimifolium, and there is a high number of species shared with all the plants studied. Overall, these results point to the central role of these understory species in the network that formed below-ground and highlight the need for awareness of their importance to the ecosystems.
Concerning Acacia longifolia, it shared symbionts with all the studied shrub species. It is possible that the shrubs play an important role by allowing the putative new fungal symbiont species to enter the network and establish themselves in the ecosystem.
These shrubs may be working as “donors” of symbionts or rather creating bridges between the already established community and the native communities by being general hosts or using other uncharacterized mechanisms.
This small-scale study revealed an appreciable diversity of ECM fungal taxa and associations in an understudied ecosystem. We found that closely related plant species showed high similarity in terms of associated fungal communities and that an invasive species has common fungal symbionts with native plants, possibly indicating that it is already part of the below-ground mutualistic network. Furthermore, a complex ECM fungal network was identified between five plant species, even in the small-scale studied area. This opens further research questions about the ecology of ECM in this coastal ecosystem. More thorough and extensive studies are needed to unravel the identity and diversity of ECM fungal species, their specificity and the ecological role of the putative CMN that connects the native species with the invasive species. A better description of how similar the communities of invasive species are in their invasive range and a comparison with their native systems will certainly provide valuable information that is needed to understand the below-ground dynamics.
A data matrix of the identified fungal taxa for each plant species can be found in Supplement 2.
The supplement related to this article is available online at: https://doi.org/10.5194/we-18-105-2018-supplement.
The authors declare that they have no conflict of interest.
We would like to thank Susana Rodriguez-Echeverria for the valuable
input in the experimental design and on data interpretation crucial for this
paper. We would like to thank Susana Gonçalves for the valuable
input through the work and expertise in the field. We would like to thank Mika Tarkka and anonymous reviewers for their valuable comments and
suggestions on an earlier version of the manuscript. This work was financed
by FCT/MEC through national funds and the co-funding by the FEDER, within
the PT2020 Partnership Agreement, and COMPETE 2020, within the Project
UID/BIA/04004/2013.
Edited by: Jutta Stadler
Reviewed by: Mika Tarkka and one anonymous referee
Agerer, R.: Exploration types of ectomycorrhizae: A proposal to classify ectomycorrhizal mycelial systems according to their patterns of differentiation and putative ecological importance, 11, 107–114, https://doi.org/10.1007/s005720100108, 2001.
Águeda, B., Parladé, J., de Miguel, A. M., and Martínez-Peña, F.: Characterization and identification of field ectomycorrhizae of Boletus edulis and Cistus ladanifer, Mycologia, 23–30, https://doi.org/10.3852/mycologia.98.1.23, 2006.
Alonso Ponce, R., Águeda, B., Ágreda, T., Modrego, M. P., Aldea, J., Fernández-toirán, L. M., and Martínez-peña, F.: Rockroses and Boletus edulis ectomycorrhizal association: Realized niche and climatic suitability in Spain, Fungal Ecol., 4, 224–232, https://doi.org/10.1016/j.funeco.2010.10.002, 2011.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 25, 3389–3402, https://doi.org/10.1093/nar/25.17.3389, 1997.
Arrington, J. M. and Kubitzki, K.: Cistaceae, in Flowering Plants ⋅ Dicotyledons, Springer, New York, USA, 62–70, 2003.
Aswathappa, N., Marcar, N. E., and Thomson, L. A. J: Salt tolerance of Australian tropical and subtropical acacias, in: Australian Acacias in Developing Countries, ACIAR Proceedings, 70–73, 1987.
Berthier, S.: Irregular Heartwood Formation in Maritime Pine (Pinus pinaster Ait): Consequences for Biomechanical and Hydraulic Tree Functioning, Ann. Bot.-London, 87, 19–25, https://doi.org/10.1006/anbo.2000.1290, 2001.
Bruns, T. D., Bidartondo, M. I., and Taylor, D. L.: Host specificity in ectomycorrhizal communities: what do the exceptions tell us?, Integr. Comp. Biol., 42, 352–359, https://doi.org/10.1093/icb/42.2.352, 2002.
Buscardo, E., Rodríguez-Echeverría, S., Martín, M. P., De Angelis, P., Pereira, J. S., and Freitas, H.: Impact of wildfire return interval on the ectomycorrhizal resistant propagules communities of a Mediterranean open forest, Fungal Biol., 114, 628–636, https://doi.org/10.1016/j.funbio.2010.05.004, 2010.
Buscardo, E., Freitas, H., Pereira, J. S., and de Angelis, P.: Common environmental factors explain both ectomycorrhizal species diversity and pine regeneration variability in a post-fire Mediterranean forest, Mycorrhiza, 21, 549–558, https://doi.org/10.1007/s00572-011-0363-5, 2011.
Buscardo, E., Rodríguez-Echeverría, S., Barrico, L., García, M. Á., Freitas, H., Martín, M. P., De Angelis, P., and Muller, L. A. H.: Is the potential for the formation of common mycorrhizal networks influenced byfire frequency?, Soil Biol. Biochem., 46, 136–144, https://doi.org/10.1016/j.soilbio.2011.12.007, 2012.
Campelo, F., Vieira, J., Battipaglia, G., de Luis, M., Nabais, C., Freitas, H., and Cherubini, P.: Which matters most for the formation of intra-annual density fluctuations in Pinus pinaster: age or size?, Trees-Struct. Funct., 29, 237–245, https://doi.org/10.1007/s00468-014-1108-9, 2015.
Chao, A., Chazdon, R. L., and Shen, T. J.: A new statistical approach for assessing similarity of species composition with incidence and abundance data, Ecol. Lett., 8, 148–159, https://doi.org/10.1111/j.1461-0248.2004.00707.x, 2005.
Comandini, O., Contu, M., and Rinaldi, A. C.: An overview of Cistus ectomycorrhizal fungi, Mycorrhiza, 16, 381–395, https://doi.org/10.1007/s00572-006-0047-8, 2006.
Cox, F., Barsoum, N., Lilleskov, E. A., and Bidartondo, M. I.: Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients, Ecol. Lett., 13, 1103–1113, https://doi.org/10.1111/j.1461-0248.2010.01494.x, 2010.
Dickie, I. A. N. A. and Reich, P. B.: Ectomycorrhizal fungal communities at forest edges, J. Ecol., 93, 244–255, https://doi.org/10.1111/j.1365-2745.2005.00977.x, 2005.
Dickie, I. A., Guza, R., and Krazewski, S.: Shared perennial ectomycorrhizal fungi between a herbacious perennial (Helianthemum bicknellii) and oak (Quercus) seedlings, New Phytol., 164, 375–382, available at: http://onlinelibrary.wiley.com/doi/10.1111/j.1469-8137.2004.01177.x/full (last access: 13 September 2017), 2004.
Dixon, P.: VEGAN, a package of R functions for community ecology, J. Veg. Sci., 14, 927–930, https://doi.org/10.1111/j.1654-1103.2003.tb02228.x, 2003.
Epskamp, S., Cramer, A. O. J., Waldorp, L. J., Schmittmann, V. D., and Borsboom, D.: qgraph?: Network Visualizations of Relationships in Psychometric Data, J. Stat. Softw., 48, https://doi.org/10.18637/jss.v048.i04, 2012.
Gamito, S.: Caution is needed when applying Margalef diversity index, Ecol. Indic., 10, 550–551, https://doi.org/10.1016/j.ecolind.2009.07.006, 2010.
Gardes, M. and Bruns, T. D.: ITS primers with enhanced specificity for basidiomycetes, application to the identification of mycorrihiza and rusts, Mol. Ecol., 2, 113–118, https://doi.org/10.1111/J.1365-294x.1993.Tb00005.X, 1993.
Godinho-Ferreira, P., Azevedo, A., and Rego, F.: Carta da Tipologia Florestal de Portugal Continental, Silva Lusit., 13, 1–34, 2005.
Guindon, S. and Gascuel, O.: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood, Syst. Biol., 52, 696–704, https://doi.org/10.1080/10635150390235520, 2003.
Horton, T. R., Cázares, E., and Bruns, T. D.: Ectomycorrhizal, vesicular-arbuscular and dark septate fungal colonization of bishop pine (Pinus muricata) seedlings in the first 5 months of growth after wildfire, Mycorrhiza, 8, 11–18, https://doi.org/10.1007/s005720050205, 1998.
Ishida, T. A., Nara, K., and Hogetsu, T.: Host effects on ectomycorrhizal fungal communities: Insight from eight host species in mixed conifer-broadleaf forests, New Phytol., 174, 430–440, https://doi.org/10.1111/j.1469-8137.2007.02016.x, 2007.
Itoo, Z. A. and Reshi, Z. A.: Effect of different nitrogen and carbon sources and concentrations on the mycelial growth of ectomycorrhizal fungi under in-vitro conditions, Scand. J. For. Res., 29, 619–628, https://doi.org/10.1080/02827581.2014.964756, 2014.
Jarvis, S., Woodward, S., Alexander, I. J., and Taylor, A. F. S.: Regional scale gradients of climate and nitrogen deposition drive variation in ectomycorrhizal fungal communities associated with native Scots pine, Glob. Change. Biol., 19, 1688–1696, https://doi.org/10.1111/gcb.12178, 2013.
Kennedy, P. G., Izzo, A. D., and Bruns, T. D.: There is high potential for the formation of common mycorrhizal networks between understorey and canopy trees in a mixed evergreen forest, J. Ecol., 91, 1071–1080, https://doi.org/10.1046/j.1365-2745.2003.00829.x, 2003.
Marchante, H., Marchante, E., and Freitas, H.: Invasion of the Portuguese dune ecosystems by the exotic species Acacia longifolia (Andrews) Willd.: effects at the community level, Plant invasions, Ecol. Threat. Manag. Solut., Backhuys Publ., Leiden, 75–85, 2003.
Marchante, H., Marchante, E., Freitas, H., and Hoffmann, J. H.: Temporal changes in the impacts on plant communities of an invasive alien tree, Acacia longifolia, Plant Ecol., 216, 1481–1498, https://doi.org/10.1007/s11258-015-0530-4, 2015.
Menkis, A., Urbina, H., James, T. Y., and Rosling, A.: Archaeorhizomyces borealis sp. nov. and a sequence-based classification of related soil fungal species, Fungal Biol., 118, 943–955, https://doi.org/10.1016/j.funbio.2014.08.005, 2014.
Monteiro-Henriques, T., Martins, M. J., Cerdeira, J. O., Silva, P., Arsénio, P., Silva, Á., Bellu, A., and Costa, J. C.: Bioclimatological mapping tackling uncertainty propagation: application to mainland Portugal, Int. J. Climatol., 36, 400–411, https://doi.org/10.1002/joc.4357, 2016.
Onnela, J. P., Saramäki, J., Kertész, J., and Kaski, K.: Intensity and coherence of motifs in weighted complex networks, Phys. Rev. E. – Stat. Nonlinear, Soft Matter Phys., 71, https://doi.org/10.1103/PhysRevE.71.065103, 2005.
Pestaña Nieto, M. and Santolamazza Carbone, S.: Characterization of juvenile maritime pine (Pinus pinaster Ait.) ectomycorrhizal fungal community using morphotyping, direct sequencing and fruitbodies sampling, Mycorrhiza, 19, 91–98, https://doi.org/10.1007/s00572-008-0207-0, 2009.
Posada, D.: jModelTest: phylogenetic model averaging, Mol. Biol. Evol., 25, 1253–1256, 2008.
Pringle, A., Bever, J. D., Gardes, M., Parrent, J. L., Rillig, M. C., and Klironomos, J. N.: Mycorrhizal Symbioses and Plant Invasions, Annu. Rev. Ecol. Evol. Syst., 40, 699–715, https://doi.org/10.1146/annurev.ecolsys.39.110707.173454, 2009.
R Development Core Team, R.: R: A Language and Environment for Statistical Computing, 2011.
Rincón, A., Santamaría-Pérez, B., Rabasa, S. G., Coince, A., Marçais, B., and Buée, M.: Compartmentalized and contrasted response of ectomycorrhizal and soil fungal communities of Scots pine forests along elevation gradients in France and Spain, Environ. Microbiol., 17, 3009–3024, https://doi.org/10.1111/1462-2920.12894, 2015.
Rodríguez-Echeverría, S.: Rhizobial hitchhikers from Down Under: Invasional meltdown in a plant-bacteria mutualism?, J. Biogeogr., 37, 1611–1622, https://doi.org/10.1111/j.1365-2699.2010.02284.x, 2010.
Rodríguez-Echeverría, S., Crisóstomo, J. A., Nabais, C., and Freitas, H.: Belowground mutualists and the invasive ability of Acacia longifolia in coastal dunes of Portugal, Biol. Invasions, 11, 651–661, https://doi.org/10.1007/s10530-008-9280-8, 2009.
Sánchez, R., Serra, F., Tárraga, J., Medina, I., Carbonell, J., Pulido, L., de María, A., Capella-Gutíerrez, S., Huerta-Cepas, J., and Gabaldón, T.: Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing, Nucleic Acids Res., 39, W470–W474, 2011.
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. a., Chen, W., Consortium, F. B., Bolchacova, E., Voigt, K., Crous, P. W., Miller, A. N., Wingfield, M. J., Aime, M. C., An, K.-D., Bai, F.-Y., Barreto, R. W., Begerow, D., Bergeron, M.-J., Blackwell, M., Boekhout, T., Bogale, M., Boonyuen, N., Burgaz, a. R., Buyck, B., Cai, L., Cai, Q., Cardinali, G., Chaverri, P., Coppins, B. J., Crespo, A., Cubas, P., Cummings, C., Damm, U., De Beer, Z. W., de Hoog, G. S., Del-Prado, R., Dentinger, B., Dieguez-Uribeondo, J., Divakar, P. K., Douglas, B., Duenas, M., Duong, T. A., Eberhardt, U., Edwards, J. E., Elshahed, M. S., Fliegerova, K., Furtado, M., Garcia, M. A., Ge, Z.-W., Griffith, G. W., Griffiths, K., Groenewald, J. Z., Groenewald, M., Grube, M., Gryzenhout, M., Guo, L.-D., Hagen, F., Hambleton, S., Hamelin, R. C., Hansen, K., Harrold, P., Heller, G., Herrera, C., Hirayama, K., Hirooka, Y., Ho, H.-M., Hoffmann, K., Hofstetter, V., Hognabba, F., Hollingsworth, P. M., Hong, S.-B. S.-B. S.-B. S.-B., Hosaka, K., Houbraken, J., Hughes, K., Huhtinen, S., Hyde, K. D., James, T., Johnson, E. M., Johnson, J. E., Johnston, P. R., Jones, E. B. G., Kelly, L. J., Kirk, P. M., Knapp, D. G., Koljalg, U., Kovacs, G. M., Kurtzman, C. P., Landvik, S., Leavitt, S. D., Liggenstoffer, A. S., Liimatainen, K., Lombard, L., Luangsa-ard, J. J., Lumbsch, H. T., Maganti, H., Maharachchikumbura, S. S. N., Martin, M. P., May, T. W., et al.: Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi, P. Natl. Acad. Sci. USA, 109, 1–6, https://doi.org/10.1073/pnas.1117018109, 2012.
Seifert, K. A.: Progress towards DNA barcoding of fungi, Mol. Ecol. Resour., 9, 83–89, https://doi.org/10.1111/j.1755-0998.2009.02635.x, 2009.
Shannon, C. E. and Weaver, W.: The Mathematical Theory of Communication, Math. theory Commun., 27, 117, https://doi.org/10.2307/3611062, 1949.
Smith, S. E. and Read, D.: Mycorrhizal Symbiosis, Academic press., Massachusetts, USA, 2006.
Taudiere, A., Munoz, F., Lesne, A., Monnet, A.-C., Bellanger, J.-M., Selosse, M.-A., Moreau, P.-A., and Richard, F.: Beyond ectomycorrhizal bipartite networks: projected networks demonstrate contrasted patterns between early- and late-successional plants in Corsica, Front. Plant Sci., 6, 1–14, https://doi.org/10.3389/fpls.2015.00881, 2015.
Tedersoo, L. and Smith, M. E.: Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground, Fungal Biol. Rev., 27, 83–99, https://doi.org/10.1016/j.fbr.2013.09.001, 2013.
Tedersoo, L., May, T. W., and Smith, M. E.: Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages, Mycorrhiza, 20, 217–263, https://doi.org/10.1007/s00572-009-0274-x, 2010.
Vrålstad, T., Myhre, E., and Schumacher, T.: Molecular diversity and phylogenetic affinities of symbiotic root-associated ascomycetes of the Helotiales in burnt and metal polluted habitats, New Phytol., 155, 131–148, https://doi.org/10.1046/j.1469-8137.2002.00444.x, 2002.
Walbert, K., Ramsfield, T. D., Ridgway, H. J., and Jones, E. E.: Ectomycorrhiza of Pinus radiata (D. Don 1836) in New Zealand – an above-and below-ground assessment, Australas. Mycol., 29, 7–16, available at: https://www.researchgate.net/publication/237091953_Ectomycorrhiza_of_Pinus_radiata_, (last access: 13 September 2017), 2010.
White, T. J., Bruns, T., Lee, S., and Taylor, J.: Amplification and direct sequencing of fungal ribossomal RNA gene for Phylogenetics – PCR Protocols, PCR Protoc., 18, 315–322, https://doi.org/10.1016/B978-0-12-372180-8.50042-1, 1990.
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y.: ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data, Methods Ecol. Evol., 8, 28–36, https://doi.org/10.1111/2041-210X.12628, 2017.
Zhang, B. and Horvath, S.: A General Framework for Weighted Gene Co-Expression Network Analysis, Stat. Appl. Genet. Mo. B., 4, https://doi.org/10.2202/1544-6115.1128, 2005.
Zunzunegui, M., Díaz Barradas, M. C., Aguilar, F., Ain Lhout, F., Clavijo, A., and García Novo, F.: Growth response of Halimium halimifolium at four sites with different soil water availability regimes in two contrasted hydrological cycles, Plant Soil, 247, 271–281, https://doi.org/10.1023/A:1021598112037, 2002.






